产品说明
一般描述
Not I recognizes the sequence GC?GG*C*CG℃ and generates fragments with 5′-cohesive termini.
Not I belongs to the class of "rare-cutter" enzymes. It is one of the two known enzymes that recognize an octameric sequence comprised solely of G and C residues.
Contents:
- Not I
- SuRE/Cut Buffer H (10x)
特异性
Recognition sites: GCGG*C*CG ℃
GCGG*C*CG ℃
Restriction site: GC↓GG*C*CG ℃
GC↓GG*C*CG ℃
Heat inactivation: Not I can be heat inactivated by incubation at 65 ℃ for 15 minutes (up to 100 U/μg DNA).
质量
Absence of nonspecific endonuclease activities
1 μg Ad2 DNA is incubated for 16 hours in 50 μl SuRE/Cut Buffer H with an excess of Not I. The number of enzyme units which do not change the enzyme-specific pattern is stated in the certificate of analysis.
Absence of exonuclease activity
Approximately 5 μg [3H] labeled calf thymus DNA are incubated with 3 μl Not I for 4 hours at +37℃ in a total volume of 100 μl 50 mM Tris-HCl, 10 mM MgCl2, 1 mM Dithioerythritol, pH approximately 7.5. Under these conditions, no release of radioactivity is detectable, as stated in the certificate of analysis.
产品规格
Average size of fragment generated
Prokaryotic genomic DNA: Not I fragments are between 20 and 1,000 kb, depending on the GC content.
Yeast genomic DNA: Not I fragments are, on average, 200 kb.
Mammalian genomic DNA: Not I fragments are approximately 1,000 kb.
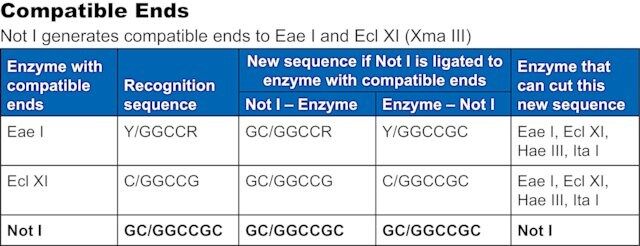
Compatible ends
Not I ends are compatible with ends generated by Eae I and EclX I (Xma III).
Isoschizomers
The enzyme has no known isoschizomers.
Methylation sensitivity
Not I is inhibited by the presence of 5-methylcytosine at the sites indicated (*) on the recognition sequence. However, the presence of 5-methylcytosine in the 5′-C position (°) is not inhibiting.
Relative activity in complete PCR mix
Relative activity in PCR mix (Taq DNA Polymerase buffer) is 0%. The PCR mix contained λDNA, primers, 10 mM Tris-HCl (pH 8.3, +20℃), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 2.5 U Taq DNA polymerase. The mix was subjected to 25 amplification cycles. After addition of 100 mM NaCl to the RE digest in the PCR mix, the activity of Not I still remains very low with below 5%.
Incubation temperature
+37℃
PFGE tested
Not I has been tested in Pulsed-Field Gel Electrophoresis (on bacterial chromosomes). For cleavage of genomic DNA (E. coli C 600) embedded in agarose for PFGE analysis, we recommend using 10 U of enzyme/μg DNA and 4 hour incubation time.
Ligation and recutting assay
Not I fragments obtained by complete digestion of 1 μg Ad2 DNA are ligated with 1 U T4 DNA Ligase in a volume of 10 μl by incubation for 16 hours at +4℃ in 66 mM Tris-HCl, 5 mM MgCl2, 5 mM Dithiothreitol, 1 mM ATP, pH 7.5 (at +20℃) resulting in >80% recovery of Ad2 DNA.
Subsequent re-cutting with Not I yields >90% of the typical pattern of Ad2 × Not I fragments.
DNA图谱分析
Number of cleavage sites on different DNAs
- λ: 0
- φX174: 0
- Ad2: 7
- M13mp7: 0
- pBR322: 0
- pBR328: 0
- pUC18: 0
- SV40: 0
单位定义
One unit is the enzyme activity that completely cleaves 1 μg Ad2DNA in one hour at +37 ℃ in a total volume of 25 μl (1x) SuRE/Cut Buffer H. The 8 fragments obtained are 18629, 6493, 5001, 2594, 1931, 954, 326 and 9 bp in length. Ad2 DNA has one Not I cleavage site that is cleaved much more slowly than the other 6 cleavage sites.
分析说明
SuRE/Cut Buffer System
The buffer in bold is recommended for optimal activity
- A: 10-25%
- B: 50-75%
- H: 100%
- L: 0-10%
- M: 25-50%
Activity in PCR buffer: 0%
其他说明
仅用于生命科学研究。不可用于诊断。
产品性质
| 质量水平 | 100 |
| 生物来源 | bacterial (Nocardia otitidis-caviarum) |
| 形式 | solution |
| 包装 | pkg of 1,000 U (11014714001 [10 U/μl]) pkg of 1,000 U (11037668001 [40 U/μl]) pkg of 200 U (11014706001 [10 U/μl]) |
| manufacturer/tradename | Roche |
| 参数 | 37 ℃ optimum reaction temp. |
| technique(s) | PCR: suitable |
| 运输 | dry ice |
| 储存温度 | −20℃ |
组份列表
组份不可单独销售
| 货号 | 组份 |
| Enzyme Solution | |
| SuRE/Cut Buffer H 10x concentrated |
安全信息
| 储存分类代码 | 12 - Non Combustible Liquids |
| WGK | WGK 1 |
| 闪点(F) | does not flash |
| 闪点(C) | does not flash |